CKDdb: a knowledge base and platform for drug development against chronic kidney disease
Introduction
Chronic Kidney Disease (CKD) is a global public health crisis, affecting an estimated 10-15% of the global population and projected to become a leading cause of death worldwide by 2040. Defined by persistent kidney damage or reduced glomerular filtration rate (GFR) for three or more months, its pathological spectrum progresses from early functional decline to irreversible glomerulosclerosis and tubulointerstitial fibrosis, culminating in end-stage kidney disease (ESKD). This progression is driven by a complex interplay of metabolic, hemodynamic, and inflammatory insults. CKD is intimately linked with extrarenal comorbidities, particularly type 2 diabetes mellitus (T2DM), cardiovascular disease (CVD), and hypertension, forming a vicious cycle of cardiorenal-metabolic derangement. While recent therapeutic advances, such as SGLT2 inhibitors and non-steroidal MRAs, have been transformative in slowing disease progression, a significant residual risk persists. Critically, no therapies can effectively reverse established fibrosis, representing a principal unmet need in nephrology.
Here we present CKDdb, a knowledge base and platform for drug development against CKD. CKDdb integrates diverse data from drug-related databases and other resources including PubMed, clinical trial registration authorities, Disease Ontology, UniProt, DrugBank, ChEMBL, TCMBank Database and CLUE. We extracted and annotated CKD-related pathogenesis (e.g., the Gut-Kidney Axis, NLRP3 Inflammasome signaling, APOL1-mediated nephropathy), in vitro & in vivo models, therapeutic strategies, targets, associated diseases and investigational drugs from published articles and clinical trial registrations as background knowledge of CKDdb. Repositioning candidates, bioactive compounds, CMap candidates and natural products were then derived from related databases as the candidate library for drug development against CKD. Drug-like properties and connectivity scores were calculated by RDKit and ConnectivityMap as the foundation of druglikeness screening and knowledge-based drug repositioning for CKD. All entities and research articles involved in CKDdb were automatically linked to associate with each other, providing knowledge graphs.
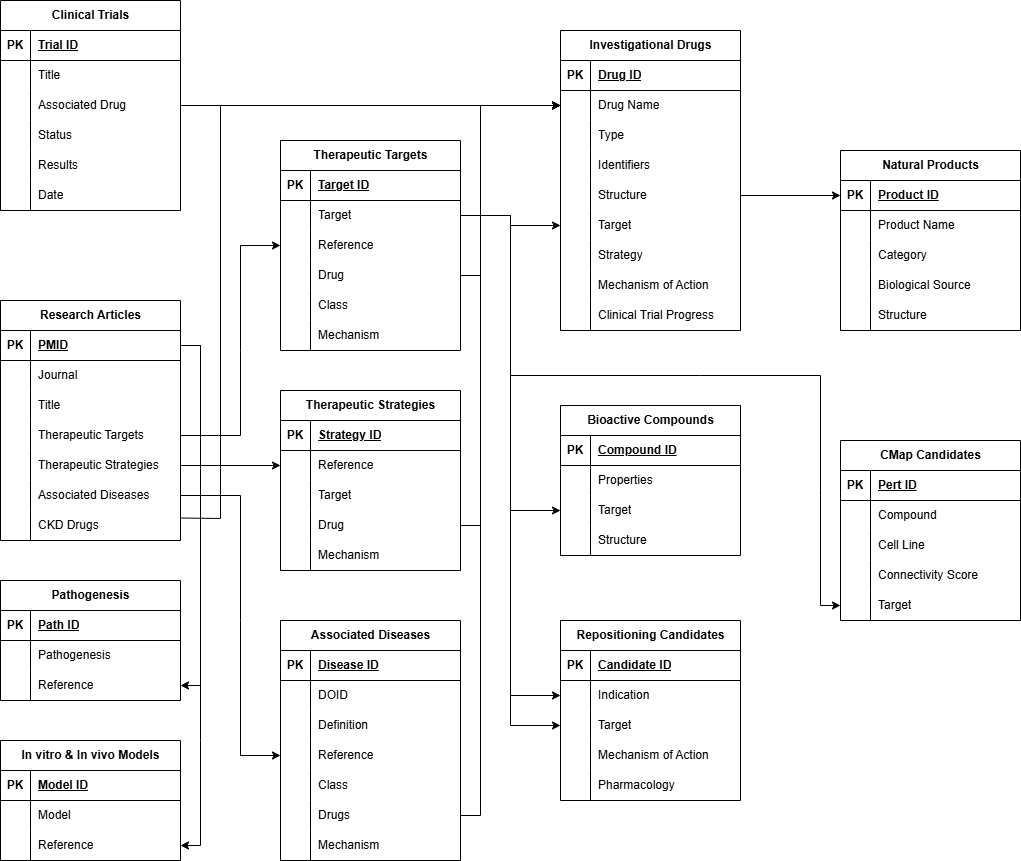
The first version of CKDdb systematically collects CKD-related therapeutic strategies, therapeutic targets, investigational drugs, research articles, repositioning candidates, bioactive compounds, CMap candidates, natural products and associated diseases from multiple public databases and other sources, all of which are freely accessible with individual detail pages and knowledge graphs. Further, a suite of tools for easier utilization and expansion is implemented, including structure search, druglikeness screening, and knowledge-based repositioning.
The ultimate goal of CKDdb is to convey the rich knowledge and provide knowledge-based analytic tools towards CKD drug development, and future updates will focus on these two aspects. To serve the wide research community, drug-related omics data, including pharmaco-genomics, -proteomics, -transcriptomics, -metabolomics, -kinetics, -dynamics data deposited in public resources will be integrated and web-based analytic tools for investigating them will be provided. Other in-depth tools for virtual screening, drug repositioning, and structure generation will also be developed. Moreover, as artificial intelligence (AI) offers powerful tools for knowledge management and bridging the gap between computational prediction and experimental validation, AI-based optimization is also expected. We will regularly update CKDdb with the latest discoveries and cutting-edge tools.
If you encounter any errors when using CKDdb, or have other feedbacks, please contact us at p2316804@mpu.edu.mo. Your comments and suggestions are welcome!